Page Not Found
Page not found. Your pixels are in another canvas.
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas.
About me
This is a page not in th emain menu
Published:
This post will show up by default. To disable scheduling of future posts, edit config.yml and set future: false.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
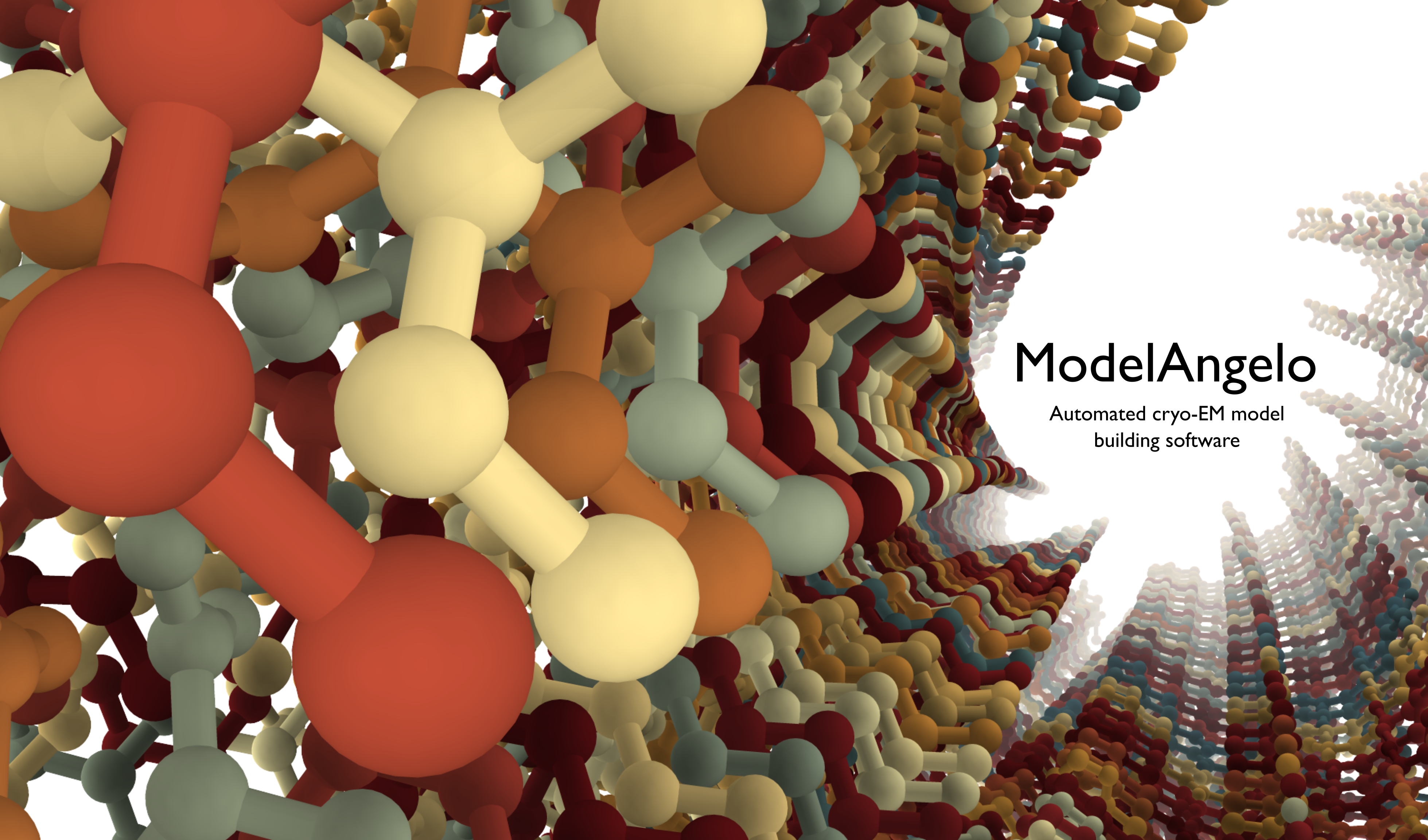
ModelAngelo is an automated cryo-EM atomic model building software. It is free and available for everyone to use on Github. You can find the pre-print on arXiv. If you have an interesting use case, and especially if you have good resolution maps with unknown protein sequences, please reach out to me! In the future, this page will include a fully worked through tutorial along with example output. For now, you can enjoy this picture of ModelAngelo’s output on PHFs.
Short description of portfolio item number 1
Short description of portfolio item number 2 
Appears on arXiv
Appears on arXiv
Published in ICLR 2020
Published in NeurIPS 2022
Published in Machine Learning for Structural Biology Workshop, NeurIPS 2022
Published in Machine Learning for Structural Biology Workshop, NeurIPS 2022
Published in ICLR 2023
Published in Nature
Published in Nature Methods
Published in ICML 2025
Published:
Gave a talk describing my PhD project on using equivariant graph neural networks to do automated model building. This was at the Swiss Equivariant Learning Workshop.
Published:
Seminar about ModelAngelo for the Molecular Biophysics Stockholm group at SciLifeLab, Stockholm.
Published:
ModelAngelo talk for the SBGrid webinar series. Recording available in the link. This talk included some practical information for running the program as well as the methods details.
Undergraduate course, University 1, Department, 2014
This is a description of a teaching experience. You can use markdown like any other post.
Workshop, University 1, Department, 2015
This is a description of a teaching experience. You can use markdown like any other post.